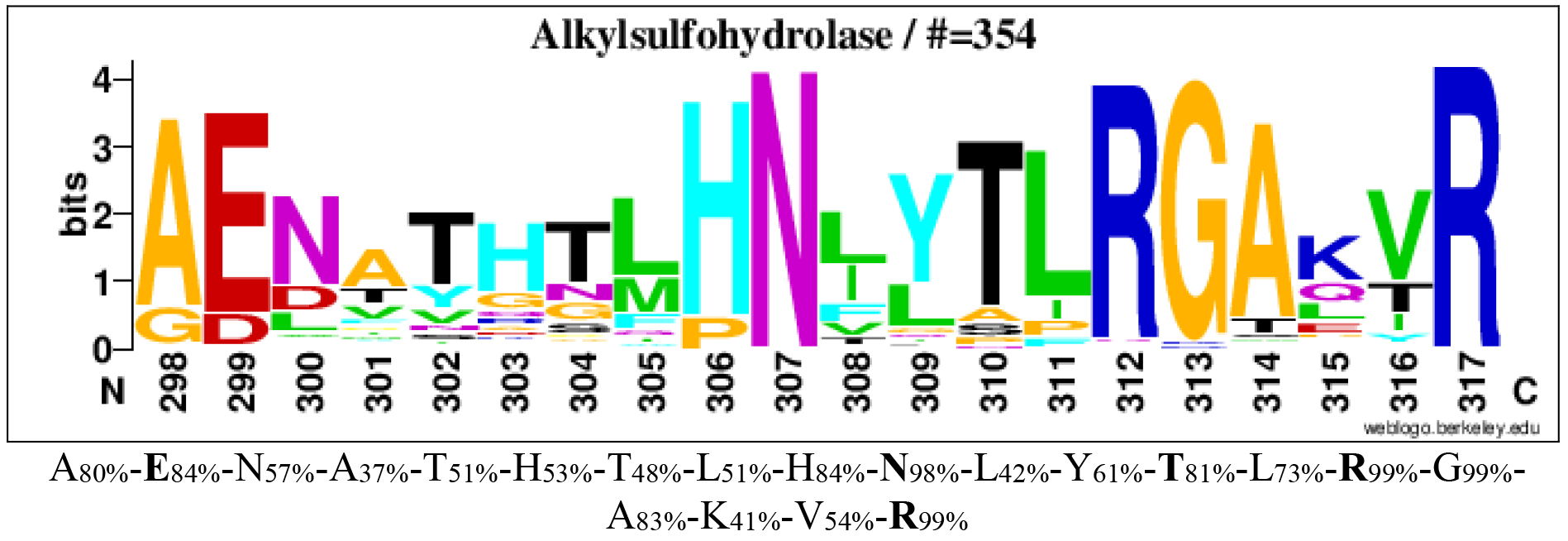Sulfatase family S3
Currently family
S3 encompasses only alkyl sulfatases. The two first characterized
family S3 enzymes, the alkylsufatases AtsK from Pseudomonas
putida S-313 (Davison et al. 1992;
Hagelueken et al. 2006) and Pisa1 from Pseudomonas
sp. DSM6611 (Knaus et al, 2012), comprise three domains: the
N-terminal catalytic domain which belongs to the
metallo-beta-lactamase superfamily and binds two zinc ions as
cofactors ; a central dimerization domain ; and a C-terminal domain
with a hydrophobic groove which was proposed to recruit long
aliphatic substrates. A conserved histidine near the sulfate-binding
site acts as the general acid for crucial protonation of the sulfate
leaving group.
Subfamilies (1)
EC activities found in subfamilies
| Subfamily |
EC Number(s) |
Description |
PubMed IDs |
The consensus sequence logoplot of family S3 (Barbeyron et al. 2016)
Logos of conserved consensus sequences identified in the global alignment of alkylsulfohydrolases (family S3).
Logos sequences identified from aligned 354 alkylsulfohydrolases. The numbers below the logo at the first position indicate the corresponding position in the reference sequence SdsA1 (Q9I5I9). The corresponding consensus sequences in multi-alignments are shown below the logo sequences. The percentages in subscript are the percentages of sequences where the amino acid is conserved in alignments. Amino acids involved in sulfate binding are in bold.
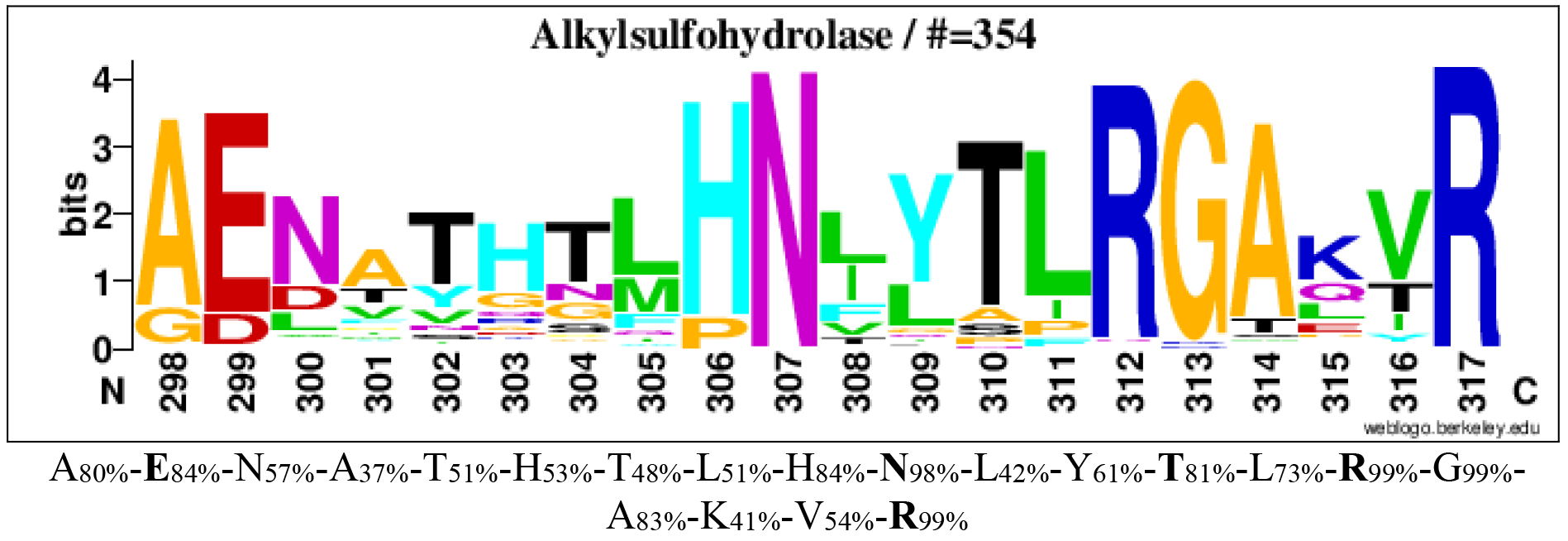
Fold and active site of a S3 family representative
Fold (A) and active site (B) of the alkylsulfatase SdsA1 from Pseudomonas aeruginosa PAO1 (PDB code: 2CFU). The fold is shown in cartoon representation. The amino acids and ligands of the active site is shown in sticks. The cations are shown as spheres. The figures were made using PyMoL (Version 1.8 Schrödinger, LLC).
 Family details
Family details